In the MOBILES project, under work package 3 (WP3), researchers are studying bacterial/microbial community in the soil to better understand how pollution affects them and how they can help restore damaged land. By analysing the genetic material of these microbes, researchers aim to identify key biological markers that indicate soil health and can guide soil rehabilitation efforts. These findings will help develop new strategies for managing contaminated environments more effectively.
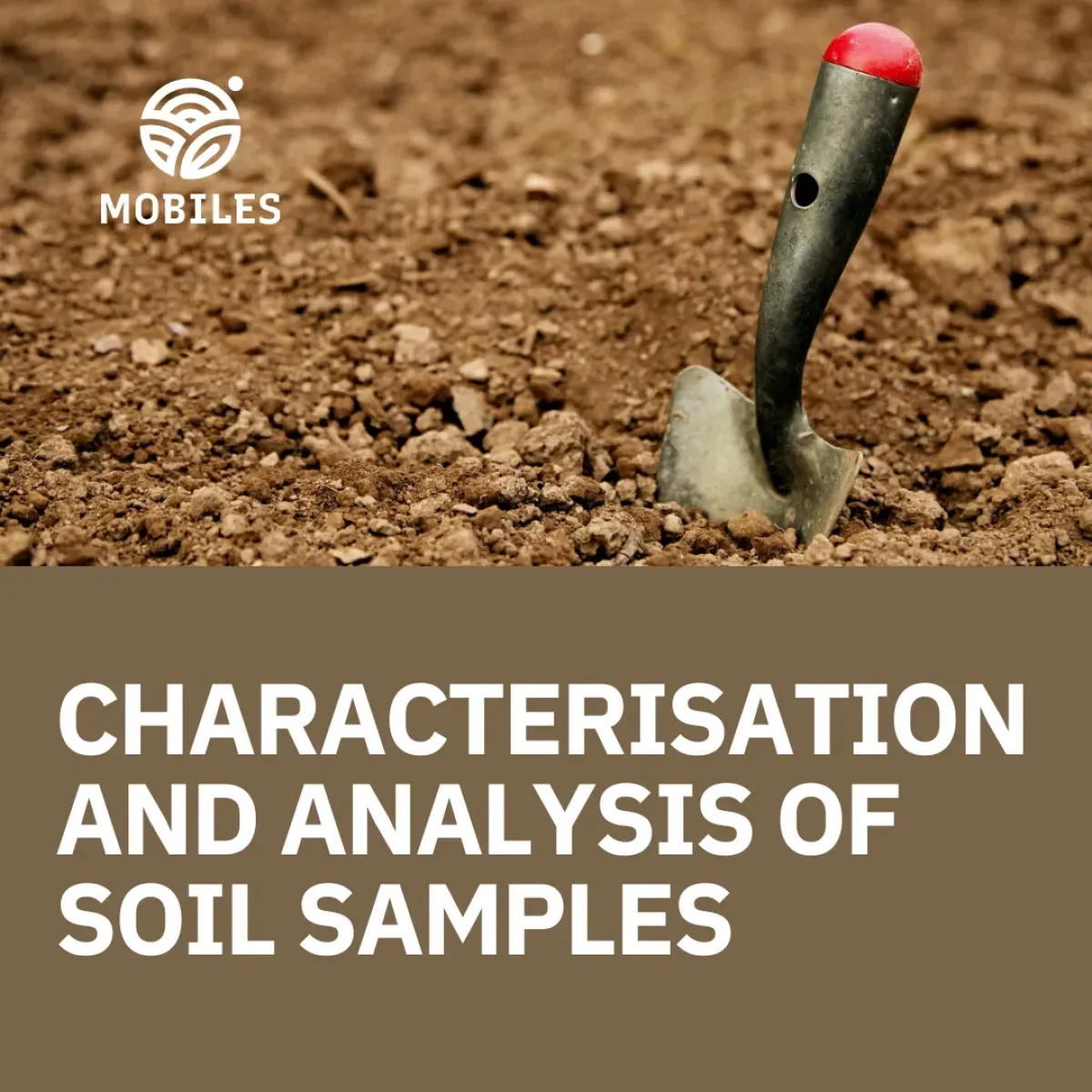
To gather data, soil samples will be taken from different locations across Europe. Sampling will be done in various seasons (spring and autumn in 2025) to see how environmental conditions influence microbial life. Collection sites will be selected in Northern and Southern Europe. Researchers have selected 6 countries, Poland, France, Italy, Germany, Greece and Cyprus (Figure 1), where they will collect soil samples; once in spring, once in autumn 2025.
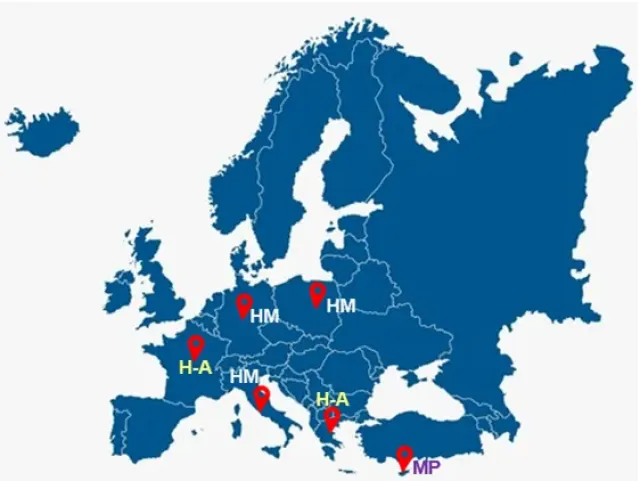
For the analyses, 3 different contamination sources were identified: Hormones, Pesticides and Antibiotics (H-A), Heavy and semi-Metals (HMs), Microplastics. A detailed protocol on the soil sampling methodology and transport conditions (Figure 2), crucial for the maintenance of RNA quality, is currently being prepared by ISSPC.
Once collected, the soil samples will be carefully delivered to the polish laboratory where, after an accurate sieving, DNA and RNA will be extracted and used for massive DNA and RNA sequencing, respectively (Figure 3). From DNA sequencing, researchers will have information on soil microbiota species composition. This, together with a chemical soil characterization, will provide critical insights on the most resilient microorganisms to a given pollutant. While, from RNA sequencing will provide important findings on the specific metabolic pathways that resilient species have evolved to survive in these harsh environments.
During the collection of soil samples, special attention will be given to soil layers up to 20 cm deep, where microbial activity is most significant. At the first stage, soil characterisation will be done for all samples evaluating: (i) pH of soil; (ii) heavy and semi-metals content characterization (As, Hg, Cd, Pb); (iii) minerals (K, P, Na, Mg, Ca, Cu); (iv) identification and possibly quantification of hormones and pesticides; (v) identification and possibly the quantification of plastic and microplastic.
Soil samples will undergo quality checks before sequencing to ensure high accuracy. Special attention will be given to microbial genes linked to pollution resistance and soil recovery, as these could be key indicators for assessing soil health and potential for rehabilitation.
Metagenomics. MOBILES project seeks to identify and understand microbial communities by DNA sequencing and taxonomical characterisation of the microbiota community in soil sample. Deeper sequencing and better reference databases are advancing the potential and success of such analyses. While meta-genomics provides information on the gene content of a microbial community and its species composition, meta-transcriptomics, using RNA extracts from soil, promises to reveal clusters identification and their actual metabolic activities at a specific time and place, and how those activities change in response to environmental forces or biotic interactions.
Researchers in the MOBILES project will use advanced sequencing technologies to study the genetic composition of soil microbes. This will help identify different species, understand their functions, and discover how they respond to pollutants.
The vast amount of genetic data collected will be processed using specific computer programs. These tools will help identify important gene clusters and patterns that could be used in future soil protection strategies. Scientists will employ bioinformatics software to compare microbial communities across different sites, tracking changes over time and under varying pollution levels. The goal is to build a comprehensive database that scientists can use to monitor and improve soil health, aiding in the development of new environmental policies and land management practices.
To store and manage all this information, researchers are creating a secure and accessible database that is already located in Spain and maintained by the University of Navarra. This system will allow scientists, beyond those involved in MOBILES project, to access and analyse the data. The infrastructure will integrate multiple data analysis tools, unified data format, data exchange mechanism, API etc. enabling researchers to visualize and interpret the results efficiently. By ensuring open access to this valuable information, the project aims to foster collaboration and drive innovation in soil conservation efforts.
The project will generate a comprehensive metagenomic database, offering valuable insights into soil microbial dynamics. The integration of sequencing, bioinformatics, and data management will provide a foundation for developing new strategies in soil protection and rehabilitation.